Hierarchical cross-entropy loss improves atlas-scale single-cell annotation models
Apr 23, 2025·,,,,,·
1 min read
Sebastiano Cultrera Di Montesano
Davide D'Ascenzo
Srivatsan Raghavan
Ava P. Amini
Peter S. Winter
Lorin Crawford
Abstract
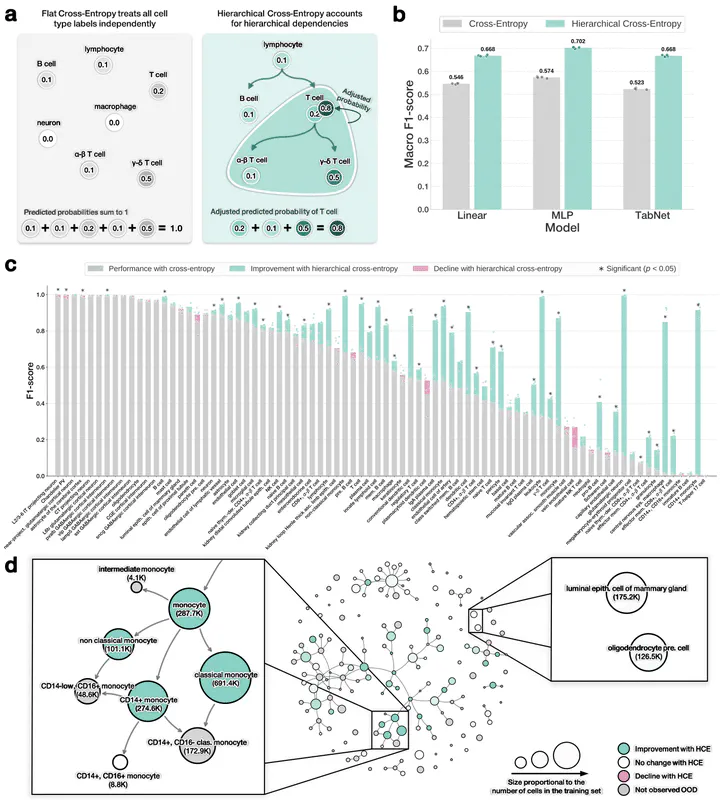
Accurately annotating cell types is essential for extracting biological insight from single-cell RNA-seq data. Although cell types are naturally organized into hierarchical ontologies, most computational models do not explicitly incorporate this structure into their training objectives. We introduce a hierarchical cross-entropy loss that aligns model objectives with biological structure. Applied to architectures ranging from linear models to transformers, this simple modification significantly improves out-of-distribution performance (12–15%) without added computational cost.
Type
Publication
bioRxiv
Accurately annotating cell types is essential for extracting biological insight from single-cell RNA-seq data. Although cell types are naturally organized into hierarchical ontologies, most computational models do not explicitly incorporate this structure into their training objectives. We introduce a hierarchical cross-entropy loss that aligns model objectives with biological structure. Applied to architectures ranging from linear models to transformers, this simple modification significantly improves out-of-distribution performance (12–15%) without added computational cost.